How to create an intensity image from a segmentation¶
from timagetk.components.labelled_image import hollow_out_labelled_image
from timagetk.io.util import shared_data
from timagetk.components.labelled_image import LabelledImage
from timagetk.visu.stack import orthogonal_view
from timagetk.io import imread
from skimage import img_as_ubyte
from timagetk.components.spatial_image import SpatialImage
from timagetk.algorithms.linearfilter import linearfilter
/builds/J-gEBwyb/0/mosaic/timagetk/src/timagetk/components/labelled_image.py:31: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from tqdm.autonotebook import tqdm
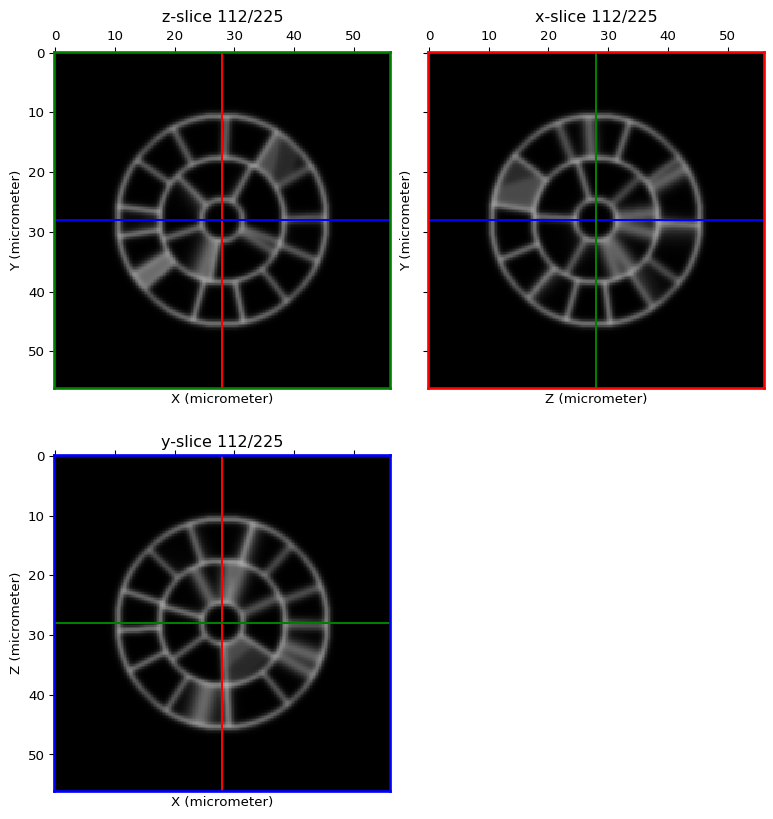
Load the example cell segmented image¶
seg_img_path = shared_data('sphere_membrane_0.0_seg.inr.gz', "sphere")
seg_img = imread(seg_img_path, rtype=LabelledImage, not_a_label=0)
orthogonal_view(seg_img, cmap='glasbey', val_range='auto')
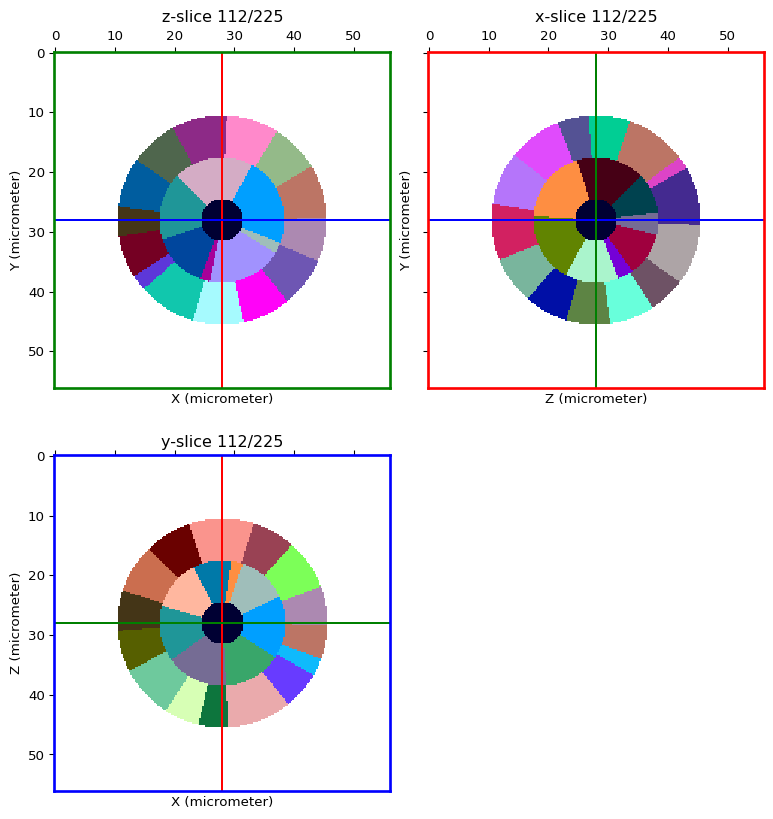
Hollow out the cells¶
hollow_seg = hollow_out_labelled_image(seg_img)
orthogonal_view(hollow_seg, cmap='glasbey', val_range='auto')
INFO [timagetk.components.labelled_image] Hollowing out labelled numpy array...
INFO [timagetk.components.labelled_image] done in 0.282s
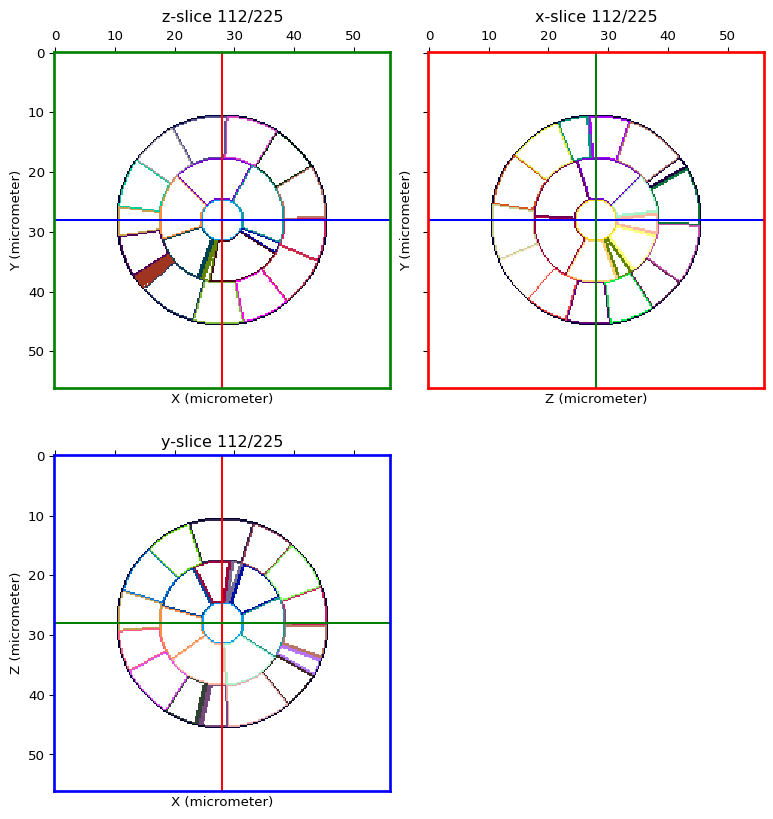
Create a membrane image from the labelled image¶
membrane_img = SpatialImage(img_as_ubyte(hollow_seg), voxelsize=seg_img.get_voxelsize()) # convert image to unsigned 8bits
membrane_img[membrane_img != 0] = 255
orthogonal_view(membrane_img, cmap='gray')
WARNING [timagetk.components.spatial_image] Undefined parameter ``axes_order``, using default: ZYX
WARNING [timagetk.components.spatial_image] Undefined parameter ``origin``, using default: [0, 0, 0]
WARNING [timagetk.components.spatial_image] Undefined parameter ``unit``, using default: 1e-06
/opt/conda/envs/timagetk/lib/python3.9/site-packages/skimage/util/dtype.py:549: UserWarning: Downcasting uint16 to uint8 without scaling because max value 64 fits in uint8
return _convert(image, np.uint8, force_copy)
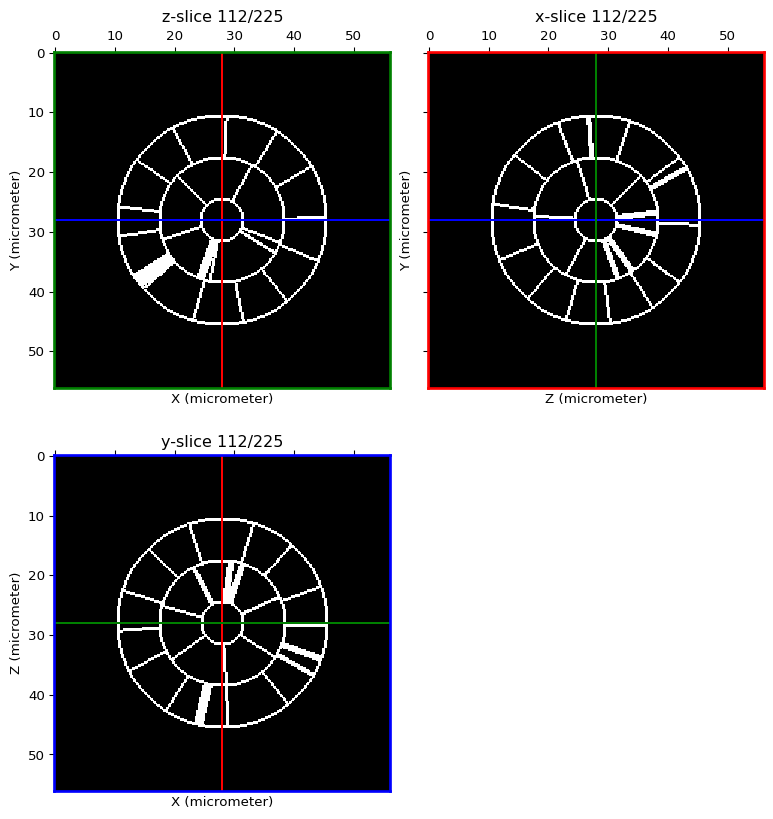
Make it a bit more realistic¶
membrane_img_real = linearfilter(membrane_img, method="smoothing", sigma=0.5, real=True)
orthogonal_view(membrane_img_real, cmap='gray')