Image conventions in computer science¶
Note
We use the widely used Python library Matplotlib to illustrate our points.
Introduction¶
To put it simply, an image is represented by a multidimensional array, and some associated metadata, e.g. the extent of the image, that is its physical size.
For example, a picture taken with your phone’s camera is not much more than three 2D arrays, one for each of the basic color: red, green and blue. Each of these arrays are filled with integers indicating the signal intensity for the corresponding color channel. Usually a picture also contains some associated metadata like when it was taken (acquisition date) or where (GPS coordinates).
For a quick example illustrating a 2D RGB image, let’s decompose our logo and view each channel separately:
import matplotlib.pyplot as plt
# - Load our PNG logo:
image = plt.imread('../_static/image/logo_timagetk.png')
# - Get the RGB channels:
red, green, blue = image[:, :, 0], image[:, :, 1], image[:, :, 2]
fig = plt.figure(figsize=(20, 5), dpi=150)
# Show original image:
fig.add_subplot(1, 4, 1)
plt.imshow(image)
plt.axis('off')
plt.title("Original RGB image")
# Show separate channels:
colors = ['Reds', 'Greens', 'Blues']
channels = [red, green, blue]
for n, (cmap, ch) in enumerate(zip(colors, channels)):
fig.add_subplot(1, 4, n+2)
plt.imshow(ch, cmap=cmap)
plt.axis('off')
plt.title(f"{cmap[:-1]} channel")
plt.show()
(Source code, png, hires.png, pdf)
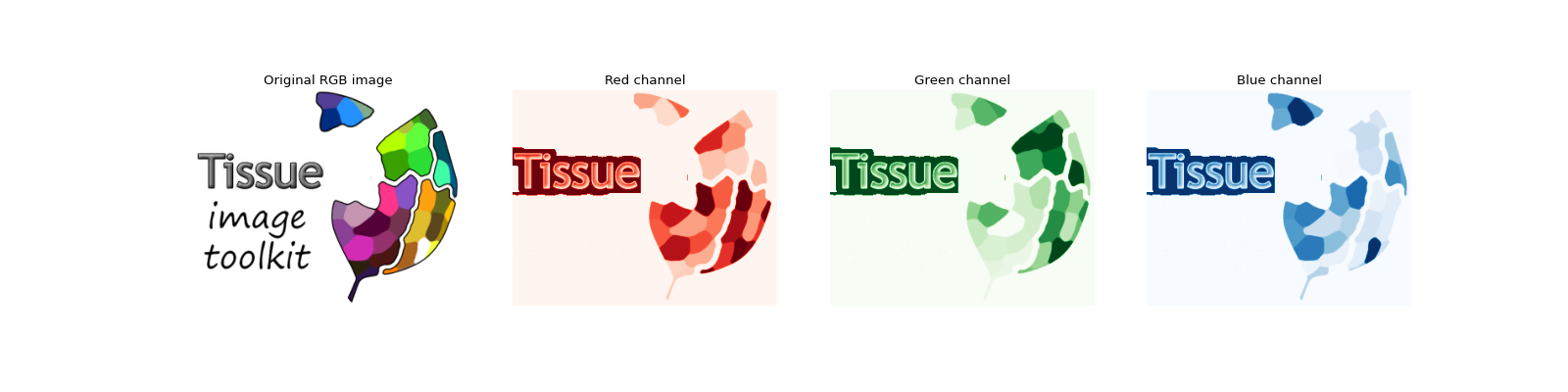
Image & coordinate conventions¶
Source: Scikit Image - Coordinate conventions
Because TimageTK uses NumPy arrays to abstract images data structure, it is important to specify the coordinate conventions. Let’s start by briefly introducing these concepts and conventions.
Cartesian coordinate system¶
In a 2D orthogonal Cartesian coordinate system, the origin is located at the bottom left corner, the abscissa x is the horizontal axis and the ordinate y the vertical one.
For more details about the Cartesian coordinate system, take a look at thisWikipedia article.
An example could be this histogram representing the distribution of a thousand randomly generated values using the numpy.random.randn() function:
(Source code, png, hires.png, pdf)

Here we can see that the hist() function from Matplotlib adhere by default to the Cartesian coordinate system conventions.
RGB image coordinate system¶
However, in a classical 2D RGB image representation, the origin is located at the top left corner, and we usually refer to the axes with the terms columns and rows rather than abscissa and ordinate. With such conventions, row refer to the vertical y-axis and column to the horizontal x-axis.
A simple illustration of this convention can be obtained using a made-up 2D array built as follows:
1. Initialize an empty (`0`) unsigned 8-bit array with max values (`255`) on the diagonal;
2. On row 3 and for all columns, set the values to `100`
It is possible to create this array using NumPy and visualize it with theimshow() function from Matplotlib:
import numpy as np
import matplotlib.pyplot as plt
# - Create a 2D unsigned 8-bit array with a diagonal at max value `255`:
arr = np.diag(np.repeat(255, 6)).astype('uint8')
# - For all columns at middle row, replace values by `100`:
arr[3, :] = 100
plt.imshow(arr, cmap='gray')
plt.colorbar()
plt.show()
(Source code, png, hires.png, pdf)

Again, we can observe that, by default, the image visualization function imshow() from matplotlib follow the convention for 2D RGB image.
Note
It is possible to change the origin location of the image. See the reference API documentation on using the origin parameter for imshow.
Microscopy image convention¶
The following schematic representation of a 2D image regroup the definition of important physical features in microscopy. This example is in 2D but generalize in 3D.

Thick indicate the row (Y) and column (X) index.
The origin is not located in the top-left corner of the top-left (origin) voxel but in the middle.
The voxel-size indicate the real size, often in µm, of the voxel
The shape of the array is
(5, 6):5for the y-axis (5 rows) and6for the x-axis (6 columns).The extent of the array is \(((\text{sh}_y - 1) * \text{vxs}_y, (\text{sh}_x - 1) * \text{vxs}_x)\), so
(4, 5)if the voxel-size is(1, 1):4for the y-axis (rows) and5for the x-axis (columns).
Multidimensional array conventions¶
The 2D image convention with 3 color channels can be extended to represent higher order images. You can indeed add planes (also called slices) to rows & columns to obtain a 3D image. Similarly, you can add a time axis to your multidimensional array to represent a dynamical image.
Important
Obviously the more axes you add to your array, the bigger the memory requirement and/or disk space!
We use the following abbreviations as array coordinates to represent image axes:
rows as
row;columns as
col;planes or slices as
pln;channels as
ch;time as
t.
It is then possible to draw to following table of image dimensions:
Image type |
Image coordinates |
Array coordinates |
Number of dimensions |
|---|---|---|---|
2D grayscale |
(row, col) |
(Y, X) |
2 |
2D multichannel |
(row, col, ch) |
(Y, X, C) |
3 |
2D time-series |
(t, row, col) |
(T, Y, X) |
3 |
2D multichannel time-series |
(t, row, col, ch) |
(T, Y, X, C) |
4 |
3D grayscale |
(pln, row, col) |
(Z, Y, X) |
3 |
3D multichannel |
(pln, row, col, ch) |
(Z, Y, X, C) |
4 |
3D time-series |
(t, pln, row, col) |
(T, Z, Y, X) |
4 |
3D multichannel time-series |
(t, pln, row, col, ch) |
(T, Z, Y, X, C) |
5 |
Important
The order of the array coordinates are important! This is not explained here, as it is a more advanced concept, but obviously it is of the upmost importance if you want to access the information!
Image encoding & data types¶
As you may know, computer use bits, i.e. 0/1, to encode & store values on disk and memory. Obviously, one bit can not hold much information, but one octet is made of 8 bits and this already allows to store larger values. This operation transforming numerical values to computer compatible values is called encoding.
Typically, an 8-bits encoding can hold the following value range:
signed: -128 to 127
unsigned: 0 to 255
Similarly, for a 16-bits encoding, the following value range will be available:
signed: -32768 to 32767
unsigned: 0 to 65535
Important
We advise to always use unsigned data types and to restrict to 8-bit or 16-bit for memory reasons.
For a more detailed explanation, refers to NumPy data type page here.
Image types¶
We hereafter define the types of images supported by timagetk and those to implement for a future release.
Important
To create an Image data structure in Python, we chose to use NumPy arrays (numpy.ndarray) to represent the signal intensity and a dictionary (dict) to organize the metadata.
Supported types¶
Low-level data structures:¶
As we have to load the image, in memory that is, we create two low-level data
The low-level image data structure is SpatialImage:
2D grayscale image (Y, X):
SpatialImage,vtImage3D grayscale image (Z, Y, X):
SpatialImage,vtImage
SpatialImage class inheritance diagram.¶
We also provide a specific data structure for segmented image:
2D labelled image (Y, X):
LabelledImage3D labelled image (Z, Y, X):
LabelledImage
LabelledImage class inheritance diagram.¶
Biology oriented data structures:¶
We also provide a specific data structure for tissue image:
2D tissue image (Y, X):
TissueImage2D3D tissue image (Z, Y, X):
TissueImage3D
TissueImage3D class inheritance diagram.¶
Fileset data structures¶
As the number of dimension grow, the memory required to create new image objects after algorithmic operation become a limitation, and we thus resort to disk access and file management.
2D/3D multi-angles
2D/3D multichannel
2D/3D time-series (unsupported yet)
2D/3D multichannel time-series (unsupported yet)
Resources¶
If you want to dig deeper into images convention in computer science we recommend the following reads:
